See ResearcherID: I-4047-2018 for full list of publications.
2025(110) RaPID discovery of cell-permeable helical peptide inhibitors con-taining cyclic β-amino acids against SARS-CoV-2 main protease. M. Kawai, T.R. Malla, H.T. Henry Chan, A. Tumber, L. Brewitz, E. Salah, N. Terasaka, T. Katoh, A. Kawamura, C.J. Schofield, F. Duarte and H. Suga. RSC Chem Biol, 2025, 6, 1089-1099 [web] (109) Macrocyclic Peptide Probes for Immunomodulatory Protein CD59: Potent Modulators of Bacterial Toxin Activity and Antibody-Dependent Cytotoxicity. J.K. Bickel, A. I. S. Ahmed, A.B. Pidd, R.M. Morgan, T. E. McAllister, S. Horrell, E. C. Couves, H. Nagaraj, E. J. Bartlett, K. E. Omari, A. Kawamura, D. Bubeck, E. W. Tate. Angewandte Chemie, 2025,137 (27), e202422673 [web] (108) Identification of selective SWI/SNF dependencies in enzalutamide-resistant prostate cancer. Gokbayrak B, Altintas UB, Lingadahalli S, Morova T, Huang CCF, Fazlioglu BE, Yu IPL, Kalkan BM, Cejas P, Kung SHY, Fazli L, Kawamura A, Long HW, Acilan C, Onder TT, Bagci-Onder T, Lynch JT, Lack NA. Communications Biology, 2025, volume 8, Article number: 169 [web] (107) Cultivating the future leaders of chemical biology. Rulka A, Adams E, Kawamura A, Wallace S. RSC Chem. Biol., 2025,6, 7-10 [web] (106) Active learning driven prioritisation of compounds from on-demand libraries targeting the SARS-CoV-2 main protease. Cree B, Bieniek MK, Amin S, Kawamura A, Cole DJ. Digital Discovery (2025). https://doi.org/10.1039/D4DD00343H [web] 2024(105) JmjC catalysed histone H2a N-methyl arginine demethylation and C4-arginine hydroxylation reveals importance of sequence-reactivity relationships. Bonnici J, Oueini R, Salah E, Johansson C, Pires E, Abboud M, Dawber RS, Tumber A, Rabe P, Saraç H, Schofield CJ, Kawamura A. Communications Biology (2024) https://doi.org/10.1038/s42003-024-07183-5 [web] (104) Active learning driven prioritisation of compounds from on-demand libraries targeting the SARS-CoV-2 main protease. Cree B, Bieniek M, Amin S, Kawamura A, Cole D. ChemRxiv (2024) https://doi.orghttps://doi.org/10.26434/chemrxiv-2024-xczfb [Web] (103) Cyclic β2,3-amino acids improve the serum stability of macrocyclic peptide inhibitors targeting the SARS-CoV-2 main protease. Miura T, Malla TR, Brewitz L, Tumber A, Salah E, Lee KJ, Terasaka N, Owen CD, Strain-Damerell C, Lukacik P, Walsh MA, Kawamura A, Schofield CJ, Katoh T, Suga H. Bulletin of the Chemical Society of Japan (2024) https://doi.org/10.1093/bulcsj/uoae018 [Web] (102) Selective targeting of human TET1 by cyclic peptide inhibitors: Insights from biochemical profiling. Šimelis K, Saraç H, Salah E, Nishio K, McAllister TE, Corner TP, Tumber A, Belle R, Schofield CJ, Suga H, Kawamura A. Bioorganic & Medicinal Chemistry (2024) https://doi.org/10.1016/j.bmc.2024.117597 [Web] (101) Identification and engineering of potent cyclic peptides with selective or promiscuous binding through biochemical profiling and bioinformatic data analysis. Smith TP, Bhaskar Bhushan B, Granata D, Kaas C, Andersen B, Decoene K, Ren Q, Liu H, Qu X, Yang Y, Pan J, Chen Q, Münzel M, Kawamura A. RSC Chemical Biology (2024) https://doi.org/10.1039/d3cb00168g [Web] (100) Focused Screening Identifies Different Sensitivities of Human TET Oxygenases to the Oncometabolite 2-Hydroxyglutarate. Belle R, Saraç H, Salah E, Bhushan B, Szykowska A, Roper G, Tumber A, Kriaucionis S, Burgess-Brown N, Schofield CJ, Brown T, Kawamura A. Journal of Medicinal Chemistry (2024) https://doi.org/10.1021/acs.jmedchem.3c01820 [Web] 2023(99) Cyclic peptides target the aromatic cage of a PHD-finger reader domain to modulate epigenetic protein function. Coleman OD, Macdonald J, Thomson B, Ward JA, Stubbs CJ, McAllister TE, Clark S, Amin S, Cao Y, Abboud MI, Zhang Y, Sanganee H, Huber KVM, Claridge TDW, Kawamura A. Chemical Science (2023) Advance Article https://doi.org/10.1039/D2SC05944D [Web] (98) In vitro selection of macrocyclic peptide inhibitors containing cyclic γ-amino acids targeting SARS-CoV-2 main protease. Miura T, Malla TR, Tumber T, Owen CD, Brewitz L, McDonough MA, Salah E, Terasaka N, Katoh T, Lukacik P, Strain-Damerell C, Mikolajek H, Walsh MA, Kawamura A, Schofield CJ, Suga H.
(97) The catalytic domains of all human KDM5 JmjC demethylases catalyse N-methyl arginine demethylation. Bonnici J, Oueini R, Salah E, Johansson C, Schofield CJ, Kawamura A. FEBS Letters (2023) 597 (7) 933-946. [Web]
2022(96) Dual LSD1 and HDAC6 inhibition induces doxorubicin sensitivity in acute myeloid leukemia cells. Bulut I, Lee A, Cevatemre B, Ruzic D, Belle R, Kawamura A, Gul S, Nikolic K, Ganesan A, Acilan C. Cancers (2022) 4(23): 6014 (95) The influence of degree of labelling upon cellular internalisation of antibody-cell penetrating peptide conjugates. Pringle TA, Coleman O, Kawamura A, Knight JC. RSC Advances (2022) 12(43), 27716-27722. [Web] This article is part of the themed collection: 2022 RSC Advances Popular Advances Collection
(94) Increasing Diversity in Admissions to Postgraduate Study. Conway SJ, Kawamura A, Marr TM, Platt F, Russell AJ. J Med Chem (2022) 65(8), 5867-5869. [Web] [PDF] (93) Reading and erasing of the phosphonium analogue of trimethyllysine by epigenetic protein. Belle R, Kamps JJAG, Poater J, Kumar K, Pieters BJGE, Salah E, Claridge TDW, Paton RS, Bickelhaupt FM, Kawamura A, Schofield CJ, Mecinović J. Commun Chem (2022) 5 27 [Web] [PDF] (92) An integrated platform approach enables discovery of potent, selective and ligand-competitive cyclic peptides targeting the GIP receptor. Bhushan B, Granata D, Kaas CS, Kasimova MA, Ren Q, Cramer CN, White MD, Hansen AMK, Fledelius C, Invernizzi G, Deibler K, Coleman OD, Zhao X, Qu X, Liu H, Zurmühl SS, Kodra JT, Kawamura A and Münzel M. Chem. Sci., (2022) 13 3256-3262 [Web] This article is part of the themed collection: Most popular 2022 organic chemistry articles
2021(91) First-in-Class Inhibitors of the Ribosomal Oxygenase MINA53. Nowak RP, Tumber A, Hendrix E, Ansari MSZ, Sabatino M, Antonini L, Andrijes R, Salah E, Mautone N, Pellegrini FR, Simelis K, Kawamura A, Johansson C, Passeri D, Pellicciari R, Ciogli A, Bufalo DD, Ragno R, Coleman ML, Trisciuoglio D, Mai A, Oppermann U, Schofield CJ, and Rotili D. J. Med. Chem (2021) 64, 23, 17031–17050 doi: https://doi.org/10.1021/acs.jmedchem.1c00605 [Web] [PDF] (90) JMJD6 is a druggable oxygenase that regulates AR-V7 expression in prostate cancer. Paschalis A, Welti J, Neeb AJ, Yuan W, Figueiredo I, Pereira R, Ferreira A, Riisnaes R, Rodrigues DN, Jiménez-Vacas JM, Kim S, Uo T, Di Micco P, Tumber T, Islam MS, Moesser MA, Abboud M, Kawamura A, Gurel B, Christova R, Gil VS, Buroni L, Crespo M, Miranda S, Lambros MB, Carreira S, Tunariu N, Alimonti A, SU2C/PCF International Prostate Cancer Dream Team, Al-Lazikani B, Schofield CJ, Plymate SR, Sharp A, de Bono JS. Cancer Res (2021) 81 (4): 1087–1100. https://doi.org/10.1158/0008-5472.CAN-20-1807 [Web] [PDF] 2020(89) The Discovery and Utility of Chemical Probes in Target Discovery (88) Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2. Chowdhury, R., Abboud, M.I., McAllister, T.E. et al. Sci Rep (2020). DOI: 10.1038/s41598-020-76307-8. [web] (87) Structural diversity in de novo cyclic peptide ligands from genetically encoded library technologies. McAllister TE, Coleman OD, Roper G, Kawamura A. Peptide Science (2020). DOI: 10.1002/pep2.24204. [web]
(86) Cyclic Peptides as Chemical Probes. Chapter 5 from The Discovery and Utility of Chemical Probes in Target Discovery, The Royal Society of Chemistry. DOI: 10.1039/9781839160745, ePub eISBN 978-1-83916-084-4 [web] (85) Light-driven post-translational installation of reactive protein side chains. Josephson B, Fehl C, Isenegger PG, Nadal S, Wright TH, Poh AWJ, Bower BJ, Giltrap AM, Chen L, Batchelor-McAuley C, Roper G, Arisa O, Sap JBI, Kawamura A, Baldwin AJ, Mohammed S, Compton RG, Gouverneur V, Davis BG. Nature (2020). doi: 10.1038/s41586-020-2733-7. [web] (84) Hypoxia and hypoxia mimetics differentially modulate histone post-translational modifications. Hsu KF, Wilkins SE, Hopkinson RJ, Sekirnik R, Flashman E, Kawamura A, McCullagh JSO, Walport L, Schofield CJ. Epigenetics DOI: 10.1080/15592294.2020.1786305 [web][PDF] (83) Engineered anti-inflammatory peptides inspired by mapping an evasin-chemokine interaction. Darlot B, Eaton JRO, Geis-Asteggiante L, Yakala GK, Karuppanan K, Davies G, Robinson CV, Kawamura A and Bhattacharya S. J Biol Chem (2020) doi: 10.1074/jbc.RA120.014103 [web][PDF]
(82) Using evasins to target the chemokine network in inflammation. Bhattacharya S & Kawamura A. Advances in Protein Chemistry and Structural Biology – Inflammatory Diseases – Part A, Volume 119, Chapter 1. Elsevier, Academic Press (ISBN: 9780128168448; Jan 2020)[web] (81) Chemical compounds targeting DNA methylation and hydroxymethylation. Belle R, Kawamura A, Arimondo PB. Epigenetics, Topics in Medicinal Chemistry (Springer Ed.) (2020) 33: 255-286. ISBN: 9783527809257 [web] (80) Inhibitors of JmjC-containing Histone Demethylases: Structural Perspective. Wright M, Brennan P, Kawamura A. Epigenetics, Topics in Medicinal Chemistry (Springer Ed.) (2020) 33: 221-253. ISBN: 9783527809257 [web] 2019 |
|||||||||||||||||||||
| (79) Systematic characterization of chromatin modifying enzymes identifies KDM3B as a critical regultor in castrate resistant prostate cancer. Saraç H, Morova T, Pires E, McCullagh J, Kaplan A, Cingöz A, Bağcı-Önder T, Önder T, Kawamura A, Lack NA. Oncogene (2019) doi:10.1038/s41388-019-1116-8 [web][PDF] | |||||||||||||||||||||
| (78) Formaldehyde quantification using ampicillin is not selective. Reinbold R, John T, Spingardi P, Kawamura A, Thompson AL, Schofield CJ, Hopkinson RJ. Scientific Report (2019) 9:18289 [web][PDF] | |||||||||||||||||||||
| (77) JmjC-domain-Containing Histone Demethylases. Højrup C, Coleman O, Bukowski JP, Clausen RP, Kawamura A. Wiley VCH, Epigenetic Drug Discovery (2018), vol 7 Methods and Principles in Medicinal Chemistry [web](76) A knottin scaffold directs the CXC-chemokine-binding specificity of tick evasins. Lee AW, Deruaz M, Lynch C, Davies G, Singh K, Alenazi Y, Eaton JRO, Kawamura A, Shaw J, Proudfoot AEI, Dias JM, Bhattacharya S. J Biol Chem (2019) 294(29) 11199–11212 [web][PDF] |
|||||||||||||||||||||
 |
|||||||||||||||||||||
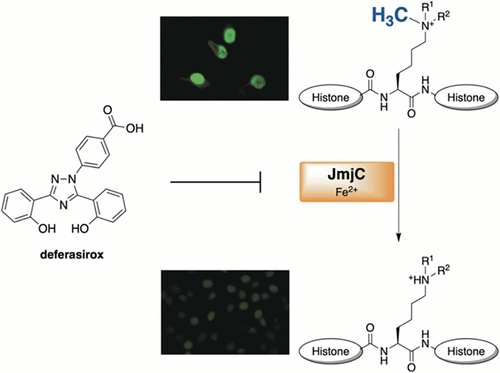
| (75) The clinically used iron chelator Deferasirox is an inhibitor of epigenetic JumonjiC domain-containing histone demethylases. Roatsch M, Hoffmann I, Abboud MI, Hancock RL, Tarhonskaya H, Hsu KH, Wilkins SE, Yeh TL, Lippl K, Serrer K, Moneke I, Ahrens TD, Robaa D, Wenzler S, Franz H, Sippl W, Lassmann S, Diederichs S, Schleicher E, Schofield CJ, Kawamura A, Schüle R, Jung M. ACS Chem. Biol. (2019) 14, 8, 1737-1750 [web][PDF] |
|||||||||||||||||||||
 |
|||||||||||||||||||||
2018 |
|||||||||||||||||||||
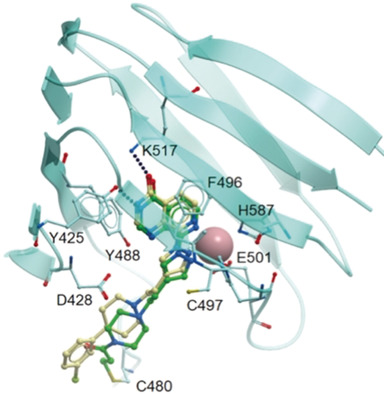
| (74) Design, Synthesis and Characterisation of Covalent KDM5 Inhibitors. Vazquez-Rodriguez S, Wright M, Rogers CM, Cribbs A, Velupillai S, Philpott M, Lee H, Dunford JE, Huber KVM, Robers MB, Vasta JD, Thezenas ML, Bonham S, Kessler B, Bennett J, Fedorov O, Raynaud F, Donovan A, Blagg J, Bavetsias V, Oppermann U, Bountra C, Kawamura A*, Brennan P*. Angew Chem Int Ed Engl 2018 58(2) 515-519 [web][PDF] Hot Paper |
|||||||||||||||||||||
 |
|||||||||||||||||||||
(73) Mechanistic and Structural Studies of KDM-Catalysed Demethylation of Histone 1 Isotype 4 at Lysine 26. Walport LJ, Hopkinson RJ, Chowdhury R, Zhang Y, Bonnici J, Schiller R, Kawamura A, Schofield CJ. (2018) FEBS Letters 592, 3264-3273 [web][PDF]. 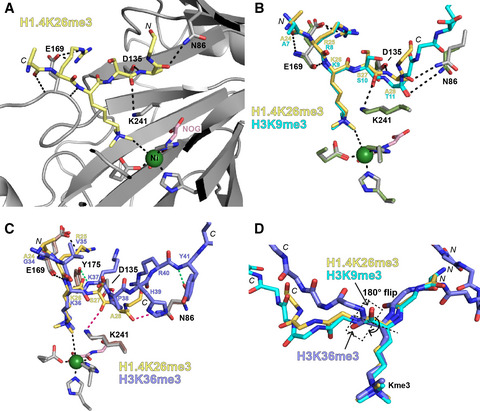 |
|||||||||||||||||||||
(72) Human Histone Demethylase KDM6B Can Catalyse Sequential Oxidations. Hopkinson RJ, Langley GW, Belle R, Walport LJ, Dunne K, Munzel M, Salah E, Kawamura A, Claridge TWD, Schofield CJ. Chem Commun (2018) 54(57): 7975–7978 [web][PDF]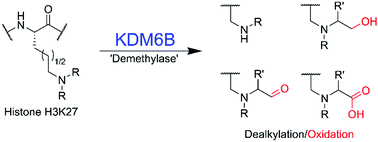 |
|
||||||||||||||||||||
(71) Synthesis and Biological Evaluation of Tripartin, a Putative KDM4 Natural Product Inhibitor, and 1-Dichloromethylinden-1-ol Analogues. Guillade L, Sarno F, Tarhonskaya T, Nebbioso A, Alvarez S, Kawamura A, Schofield CJ, Altucci L, de Lera AR. ChemBioChem (2018) 13, 1-9 [web][PDF]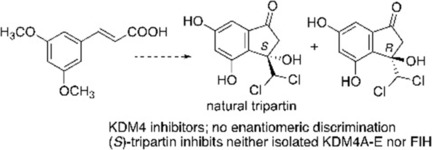 |
|
||||||||||||||||||||
(70) Non-competitive Cyclic Peptides for Targeting Enzyme-substrate Complexes. McAllister T, Zhang TL, Abboud MI, Leung KH, Hookway ES, King ONF, Bhushan B, Williams ST, Hopkinson RJ, Munzel M, Loik ND, Chowdhury R, Oppermann U, Claridge TDW, Goto Y, Suga H, Schofield CJ, Kawamura A*. Chemical Science (2018) 9, 4569 – 4578 [web][PDF]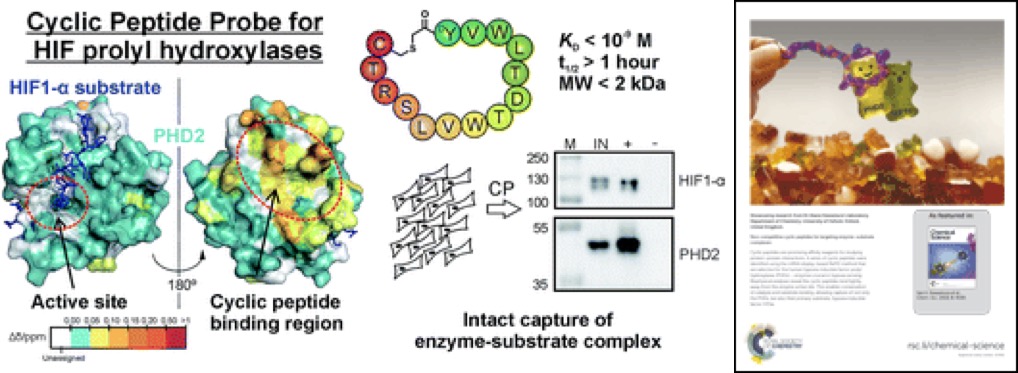 |
|
||||||||||||||||||||
| (69) Genetically engineered two-warhead chemokine binding proteins provide a method to achieve precision targeting of disease-relevant chemokine subsets. Alenazi Y, Singh K, Daives G, Eaton JRO, Kawamura A & Bhattacharya S. Scientific Reports (2018), Article number: 6333 [web][PDF] | |||||||||||||||||||||
(68) Investigations on small molecule inhibitors targeting the histone H3K4 tri-methyllysine binding PHD-finger of JmjC histone demethylases. Bhushan B, Erdmann A, Yijia Zhang, Belle R, Johannson C, Oppermann U, Hopkinson RJ, Schofield CJ, Kawamura A*. Bioorg Med Chem, 26(11):2984-2991 [web][PDF]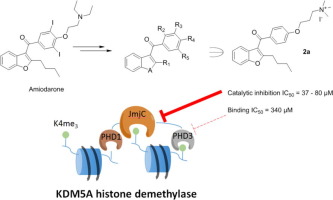 |
|
||||||||||||||||||||
| (67) Functional characterization of a novel tick CC chemokine binding protein identifies a transportable CCL8 binding domain. Eaton JRO, Alenazi Y, Singh K, Davies G, Geis-Asteggiante L, Kessler B, Robinson CV, Kawamura A, Bhattacharya S. Journal of Biological Chemistry (2018) 293, 6134-6146 [web][PDF] | |||||||||||||||||||||
(66) Thioether Macrocyclic Peptides Selected against TET1 Compact Catalytic Domain Inhibit TET1 Catalytic Activity. Nishio K, Belle R, Katoh T, Kawamura A, Sengoku T, Hanada K, Ohsawa N, Shirouzu M, Yokoyama S, Suga H. ChemBioChem (2018), 19(9):979-985 [web][PDF] VIP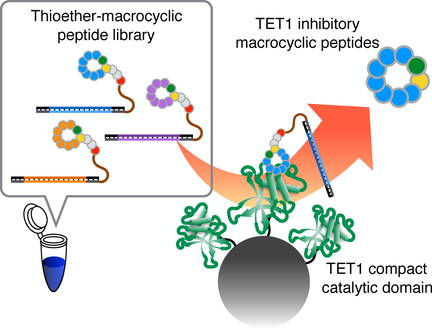 |
|
||||||||||||||||||||
(65) Lysine-241 has a role in coupling 2OG with substrate oxidation during KDM4A-catalysed histone demethylation. Hancock RL, Smart TJ, Flashman E, Kawamura A, Schofield CJ, Hopkinson R. ChemBioChem (2018) 19(9):917-921 [web][PDF] VIP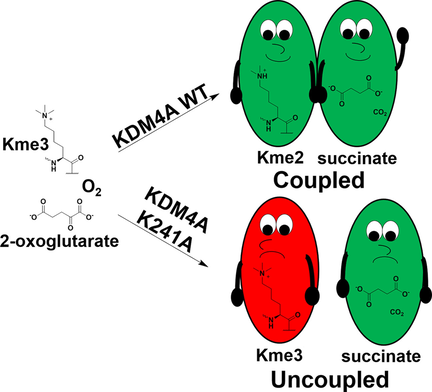 |
|
||||||||||||||||||||
(64) Structure-activity studies of a macrocyclic peptide inhibitor of histone lysine demethylase 4A. Passioura T, Bhushan B, Tumber A, Kawamura A*, Suga H*. Bioorg Med Chem, 26(6):1225-1231 [web][PDF]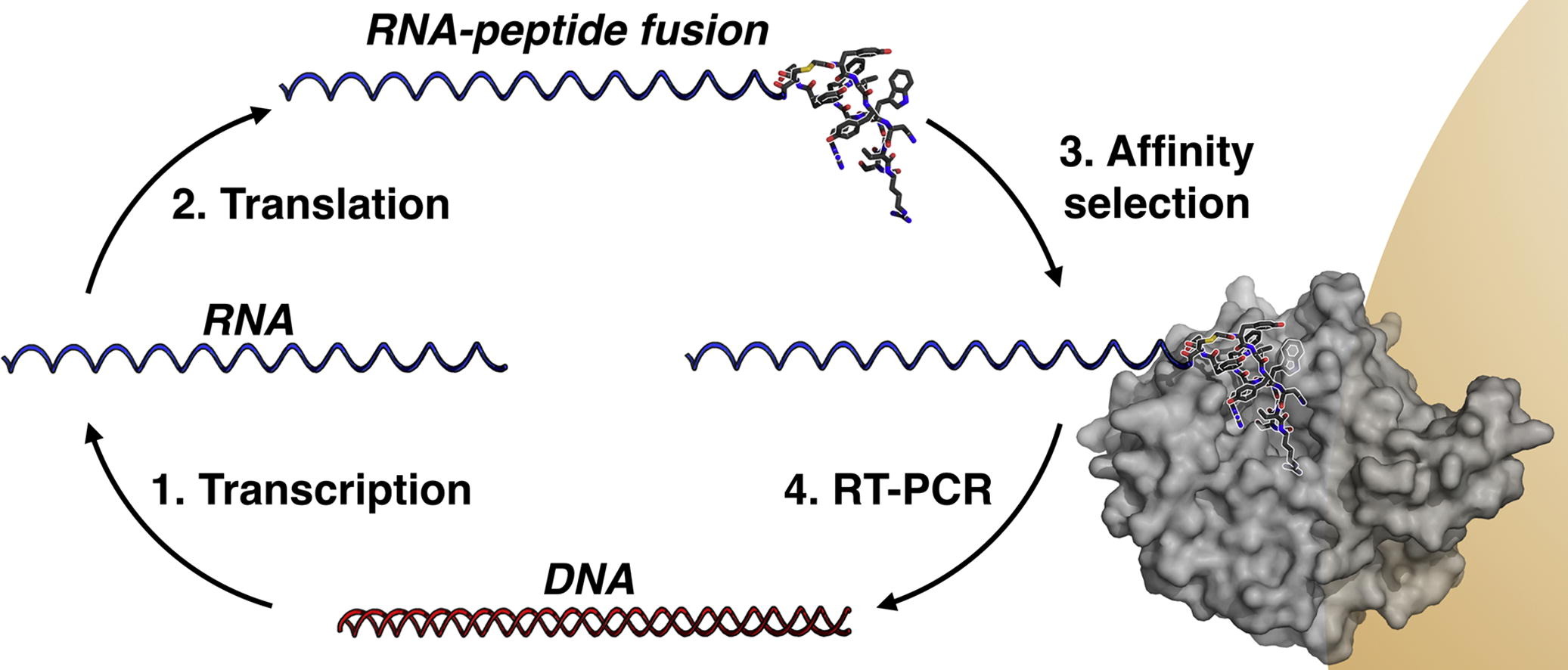 |
|
||||||||||||||||||||
(63) 2-Oxoglutarate Regulates Binding of Hydroxylated Hypoxia-Inducible Factor to Prolyl Hydroxylase Domain 2. Abboud MI, McAllister T, Leung IKH, Chowdhury R, Jorgensen C, Domene C, Mecinović J, Lippl K, Hopkinson RJ, Kawamura A, Claridge TDW, Schofield CJ. ChemBioChem (2018) 54(25):3130-3133 [web][PDF]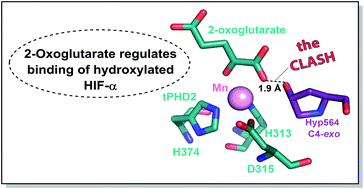 |
|
||||||||||||||||||||
| (62) In Vitro Enzyme Assays for Lysine Histone Demethylases. Tarhonskaya H, Tumber A, Kawamura A*, Schofield CJ*. Current Protocols in Pharmacology (2018) 80(1):3.15.1-3.15.12. [web] | |||||||||||||||||||||
| (61) Inhibitors of both the N-methyl lysyl- and arginyl- demethylase activities of the JmjC oxygenases. Bonnici J, Tumber A, Kawamura A, Schofield CJ. Royal Society Philosophical Transactions B (2018) 373(1748). pii: 20170071 [web][PDF] | |||||||||||||||||||||
2017 |
|||||||||||||||||||||
(60) Investigating D-Lysine Stereochemistry for Epigenetic Methylation, Demethylation and Recognition. Belle R, Temimi AHKA, Pieters BJGE, Kumar K, Tumber T, Dunford JE, Johansson C, Oppermann U, Brown T, Schofield CJ, Hopkinson RJ, Paton RS, Kawamura A, Mecinović J. Chemical Communications (2017) 53(99), 13264-13267 [web][PDF] 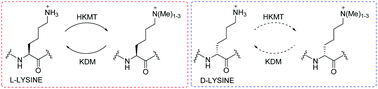 |
|
||||||||||||||||||||
(59) Discovery of a highly selective cell-active inhibitor of KDM2/7. Gerken PA, Wolstenhulme JR, Tumber A, Hatch SB, Zhang Y, Müller S, Chandler SA, Mair B, Li F, Nijman SBM, Konietzny R, Szommer T, Fedorov O, Benesch JLP, Vedadi M, Kessler BM, Kawamura A, Brennan PE, Smith ME. Angewandte Chemie Intl Ed (2017) 56(49):15555-15559 [web][PDF] 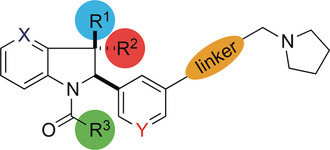 |
|
||||||||||||||||||||
(58) Molecular and Cellular Mechanisms of HIF Prolyl Hydroxylase inhibitors in Clinical Trials. Yeh TL, Leissing TM, Abboud MI, Thinnes CT, Atasoylu O, Holt-Martyn JP, Zhang D, Tumber A, Lippl K, Lohans CT, Leung IKH, Morcrette H, Kawamura A, Flashman E, Lu X, Ratcliffe PJ, Chowdhury R, Pugh CW, Schofield CJ. Chemical Science (2017), 429, 2895-2906 [web][PDF] 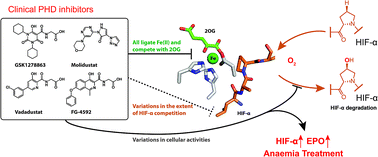 |
|
||||||||||||||||||||
| (57) Role of KDM4A histone demethylase in HIF-1α activity in hypoxia. Dobrynin G, Ramachandran S, McAllister T, Kawamura A and Hammond EM. Scientific Reports (2017), Article number: 11094 [web][PDF] | |||||||||||||||||||||
(56) Studies on the Interaction of the Histone Demethylase KDM5B with Tricarboxylic Acid Cycle Intermediates. Tarhonskaya H, Nowak RP, Johansson C, Szykowska A, Tumber A, Hancock RL, Lang P, Flashman F, Oppermann U, Schofield CJ* and Kawamura A*. Journal of Molecular Biology (2017), 429(19):2895-2906 [web][PDF] 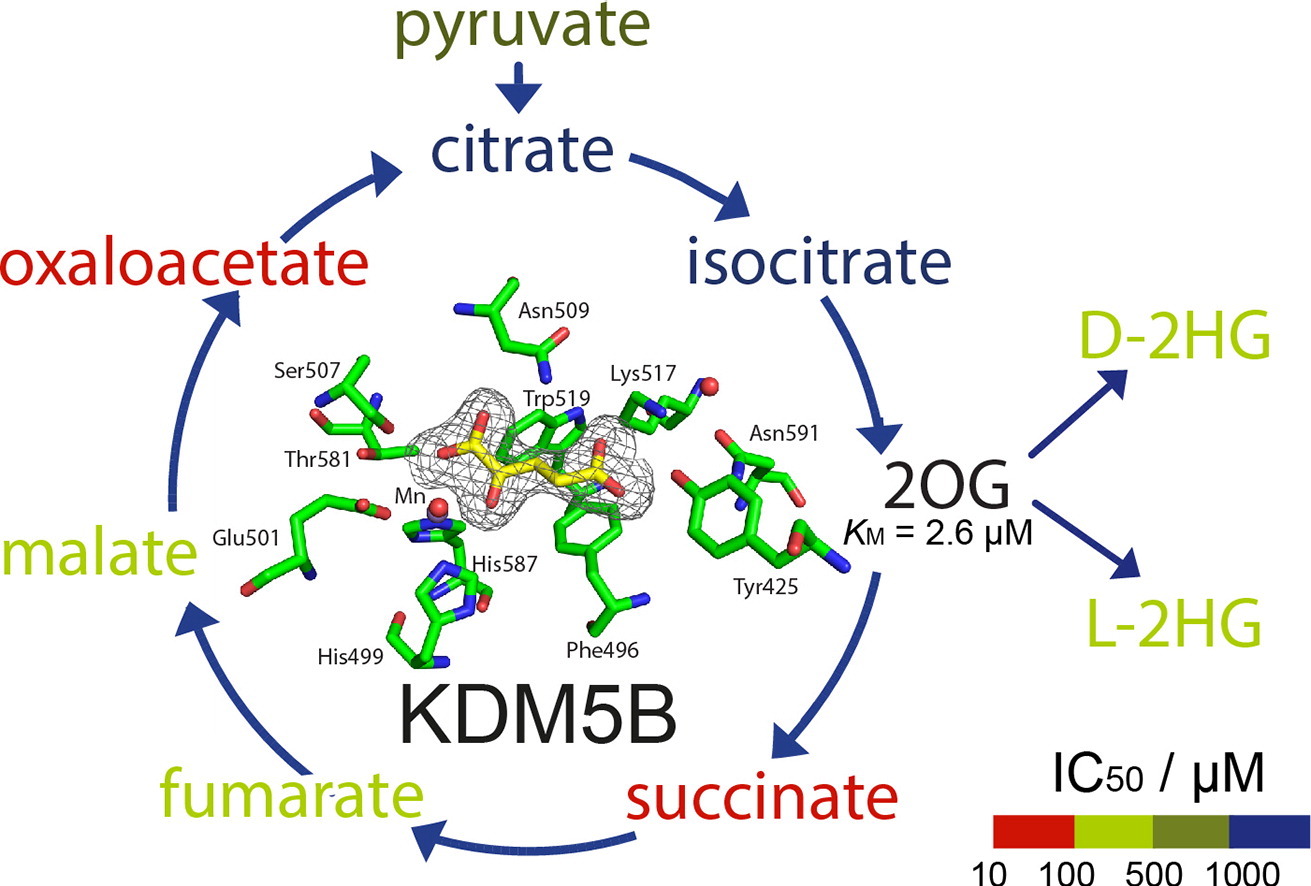 |
|
||||||||||||||||||||
| (55) Yeast surface display identifies a novel family of polyvalent CC chemokine-binding evasin peptides from ticks. Singh K, Graham D, Alenazi Y, Eaton J, Kawamura A, Bhattacharya S. Scientific Reports (2017) 7: 4267 [web][PDF] Featured on the BBC | |||||||||||||||||||||
| (54) Highly Selective Inhibition of Histone Demethylases by De Novo Macrocyclic Peptides. Kawamura A*, Münzel M, Kojima T, Yapp C, Bhushan B, Goto Y, Tumber A, Katoh K, King ONF, Passioura T, Walport LJ, Hatch SB, Madden M, Müller M, Brennan PE, Chowdhury R, Hopkinson RJ, Suga H* & Schofield CJ* Nature Communications (2017) 8:14773 [web][PDF] | |||||||||||||||||||||
(53) The activity of JmjC histone lysine demethylase KDM4A is highly sensitive to oxygen concentrations. Hancock RL, Mason N, Dunne K, Flashman E*, Kawamura A*. ACS Chemical Biology (2017) 12(4):1011-1019 [web][PDF]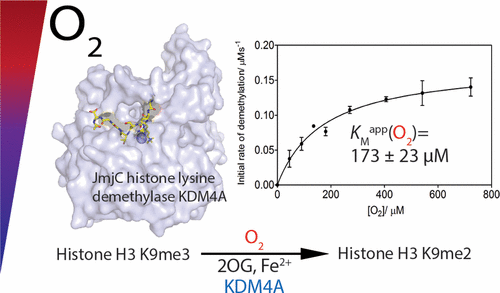 |
|
||||||||||||||||||||
| (52) Assessing Histone Demethylase Inhibitors in Cells – Lessons Learned. Hatch SB, Yapp C, Montenegro RC, Savitsky P, Gamble V, Tumber A, Bavetsias V, Fedorov O, Nuzzi A, Atrash B, Raynaud F, Lanigan R, Rossanese O, Westaway SM, Brown JA, Prinjha RK, Oppermann U, Schofield CJ, Bountra C, Kawamura A, Blagg J, Brennan PE, Müller S. Epigenetics & Chromatin (2017) 10:9. doi: 10.1186/s13072-017-0116-6 [web][PDF] | |||||||||||||||||||||
(51) Selective recognition of the di/trimethylammonium motif by an artificial carboxycalixarene receptor. Hanauer T, Hopkinson RJ, Patel K, Li Y, Correddu D, Kawamura A, Sarojini V, Leung IKH, Gruber T. Org. Biomol. Chem (2017) 15 (5), 1100-1105 [web][PDF]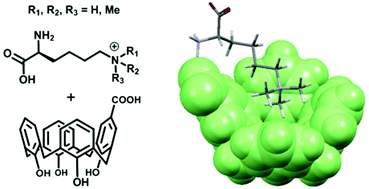 |
|
||||||||||||||||||||
| (50) KDM3A coordinates actin dynamics with intraflagellar transport to regulate cilia stability. Yeyati P, Schiller R, Mali G, Kasioulis I, Kawamura A, Adams IR, Playfoot CJ, Gilbert N, van Heyningen V, Wills J, von Kriegsheim A, Finch A, Sakai J, Jackson I, Schofield CJ, and Mill P. Journal of Cell Biology (2017) doi: 10.1083/jcb.201607032. [web][PDF] | |||||||||||||||||||||
(49) A Selective KDM5 Inhibitor, KDOAM-25, inhibits proliferation of MM1S myeloma cells in vitro. Tumber A, Nuzzi A, Hookway E, Hatch S, Velupillai S, Kawamura A, Savitsky P, Yapp C, Szykowska A, Wu N, Johansson C, Bountra C, Strain-Damerell C, Burgess-Brown N, Ruda GF, Fedorov O, Munro S, England KS, Schofield CJ, LaThangue N, Pawlyn C, Davies F, Morgan G, Athanasou N, Müller S, Oppermann U, Brennan PE. Cell Chemical Biology (2017) 24(3):371-380 [web][PDF]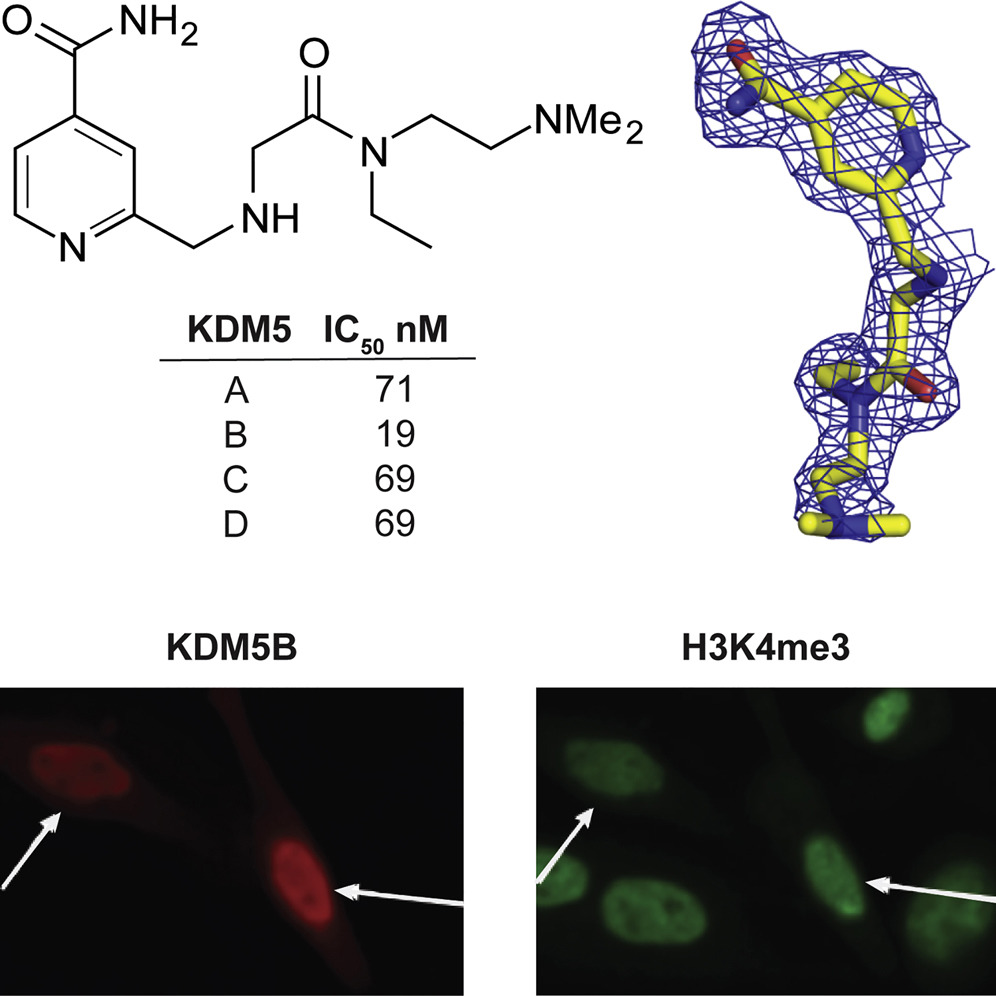 |
|
||||||||||||||||||||
2016
|
|||||||||||||||||||||
| (48) Arginine demethylation is catalyzed by a subset of JmjC histone lysine demethylases. Walport LJ, Hopkinson RJ, Schiller R, Chowdhury R, Kawamura A, Schofield CJ. Nature Communications (2016) 7, 11974 [web][PDF] | |||||||||||||||||||||
(47) Tetracyanoresorcin[4]arene selectively recognises trimethyllysine and inhibits its enzyme-catalysed demethylation. Peacock H, Thinnes C, Kawamura A, Hamilton A. Supramolecular Chemistry (2016) 28 575-581 [web][PDF]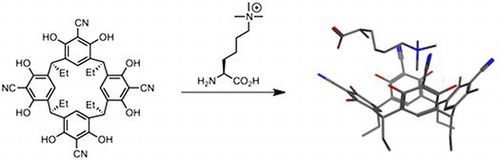 |
|
||||||||||||||||||||
(46) 8-Substituted pyrido[3,4-d]pyrimidin-4(3H)-one derivatives as potent, cell permeable, KDM4 (JMJD2) and KDM5 (JARID1) histone demethylase inhibitors. Bavetsias V, Lanigan RM, Ruda GF, Atrash B, McLaughlin MG, Tumber A, Mok NY, LeBihan YV, Dempster S, Boxall KJ, Jeganathan F, Velupilliai S, Krojer T, England KS, Sejberg J, Thai C, Donovan A, Pal A, Scozzafava G, Bennett JM, Kawamura A, Johansson C, Szykowska A, Gileadi C, Burgess-Brown NA, von Delft F, Oppermann U, Walters Z, Shipley J, Raynaud FI, Westaway SM, Prinjha RK, Fedorov O, Burke R, Schofield CJ, Westwood IM, Bountra C, van Montfort RLM, Brennan P, Blagg J. J Med Chem (2016) 59(4):1388-409 [web][PDF]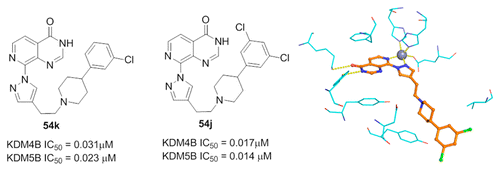 |
|
||||||||||||||||||||
(45) Recent Progress in Histone Demethylase Inhibitors. McAllister TE, England KS, Hopkinson RJ, Brennan PE, Kawamura A*, Schofield CJ*. J Med Chem (2016) 59(4):1308-29 [web][PDF]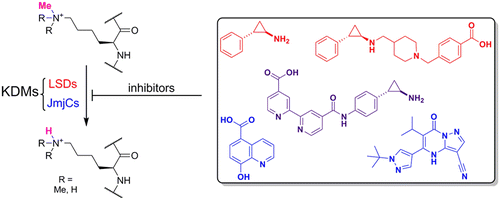 |
|
||||||||||||||||||||
2015
|
|||||||||||||||||||||
(44) Betti reaction enables efficient synthesis of 8-hydroxyquinoline inhibitors of 2-oxoglutarate oxygenases. Thinnes CC, Tumber A, Yapp C, Scozzafava G, Yeh T, Chan MC, Tran TA, Hsu K, Tarhonskaya H, Walport LJ, Wilkins SE, Martinez ED, Müller S, Pugh CW, Ratcliffe PJ, Brennan PE, Kawamura A*, Schofield CJ*. Chem Commun (Camb) (2015) 51(84):15458-61 [web][PDF]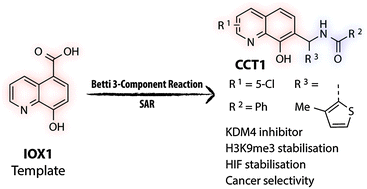 |
|
||||||||||||||||||||
| (43) Potent and Selective Triazole-based Inhibitors of the Hypoxia-inducible Factor Prolyl-hydroxylases with Activity in the Murine Brain. Chan MC, Atasoylu O, Hodson E, Tumber A, Leung IKH, Chowdhury R, Gómez-Pérez V, Demetriades M, Rydzik AM, Holt-Martyn J, Tian YM, Bishop T, Claridge TDW, Kawamura A, Pugh CW, Ratcliffe PJ, Schofield CJ. PLoS ONE (2015) Jul 6;10(7):e0132004 [web][PDF] | |||||||||||||||||||||
2014 |
|||||||||||||||||||||
(40) Select this element Rhodanine hydrolysis leads to potent thioenolate mediated metallo-β-lactamase inhibition. Brem J, van Berkel SS, Aik W, Rydzik AM, Avison MB, Pettinati I, Umland KD, Kawamura A, Spencer J, Claridge TD, McDonough MA, Schofield CJ. Nature Chemistry (2014) 6(12):1084-1090 [web][PDF] |
|
||||||||||||||||||||
| (39) Differences between murine arylamine N-acetyltransferase Type 1 and human arylamine N-acetyltransferase Type 2 defined by substrate specificity and inhibitor binding. Laurieri N, Kawamura A, Westwood IM, Varney A, Morris E, Russell AJ, Stanley LA and Sim E. BMC Pharmacology and Toxicology (2014) 15:68 [web][PDF]. 10.1186/2050-6511-15-68 | |||||||||||||||||||||
(38) Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor. England KS, Tumber A, Krojer T, Scozzafava G, Ng SS, Daniel M, Szykowska A, Che KH, von Delft F, Burgess-Brown NA, Kawamura A, Schofield CJ, Brennan PE. Med. Chem. Commun. (2014), 5, 1879-1886 [web][PDF] |
|
||||||||||||||||||||
(37) Modulating carnitine levels by targeting its biosynthesis – selective inhibition of γ-butyrobetaine hydroxylase. Rydzik AM, Chowdhury R, Kochan GT, Williams ST, McDonough MA, Kawamura A, Schofield CJ. Chemical Science (2014), 5, 1765-1771 [web][PDF] |
|
||||||||||||||||||||
| (36) A Cell-Permeable Ester Derivative of the JmjC Histone Demethylase Inhibitor IOX1. Schiller R, Wickens JR, Bush JT, Lejeune C, Choi H, Yeh T-L, Chan MC, Mccullagh JSO, Schofield CJ*, Kawamura A*. Chem. Med. Chem. (2014) 9(3):566-71 [web][PDF] | |||||||||||||||||||||
| (35) Non-enzymatic chemistry enables 2-hydroxyglutarate-mediated activation of 2-oxoglutarate oxygenases. Tarhonskaya H, Rydzik AM, Leung IK, Loik ND, Chan MC, Kawamura A, McCullagh JS, Claridge TD, Flashman E, Schofield CJ. Nature Communications (2014) 5:3423 [web][PDF]
|
|||||||||||||||||||||
| (34) Targeting histone lysine demethylases – Progress, challenges, and the future. Thinnes CC, England KS, Kawamura A, Chowdhury R, Schofield CJ, Hopkinson RJ. Biochim Biophys Acta. (2014). pii: S1874-9399(14)00115-1 [web][PDF] | |||||||||||||||||||||
(33) Pan-Histone Demethylase Inhibitors Simultaneously Targeting Jumonji C and Lysine Specific Demethylases Display High Anticancer Activities. Rotili D, Tomassi S, Conte M, Benedetti R, Tortorici M, Ciossani G, Valente S, Marrocco B, Labella D, Novellino E, Mattevi A, Altucci L, Tumber A, Yapp C, King ON, Hopkinson RJ, Kawamura A, Schofield CJ, Mai A. J Med Chem (2014) 57(1):42-55 [web][PDF] |
|
||||||||||||||||||||
2013 |
|||||||||||||||||||||
(32) 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation. Hokpkinson RJ, Tumber A, Yapp C, Chowdhury R, Aik W, Che KH, Li XL, Kristensen JBL, King ON, Chan MC, Yeoh KK, Choi H, Walport LJ, Thinnes CC, Bush JT, Lejeune C, Rydzik AM, Rose NR, Bagg EA, Donough MA, Yue WW, Ng SS, Olse L, Brennan P, Oppermann U, Muller-Knapp S, Klose RJ, Ratcliffe PJ, Schofield CJ*, Kawamura A* Chemical Science (2013) 4, 3110-3117 [web][PDF]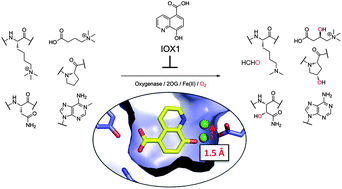 |
|
||||||||||||||||||||
(31) Identification of the KDM2/7 Histone Lysine Demethylase Subfamily Inhibitor and its Antiproliferative Activity. Suzuki T, Ozasa H, Itoh Y, Mino K, Walport L, Ohkubo R, Kawamura A, Yonezawa M, Tsukada Y, Tumber A, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Schofield CJ, Miyata N. J. Med. Chem. (2013) 56(18):7222-7231 [web][PDF]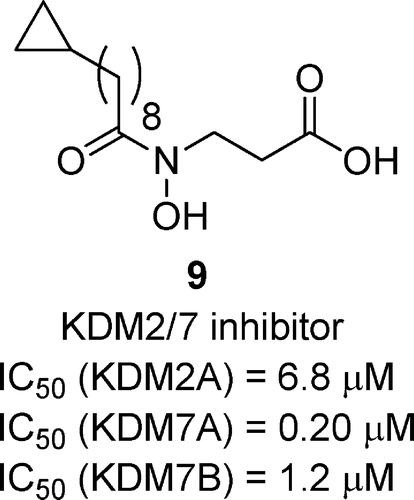 |
|
||||||||||||||||||||
(30) Is JmjC Oxygenase Catalysis Limited to Demethylation? Hopkinson RJ, Walport LJ, Münzel M, Rose NR, Smart TJ, Kawamura A, Claridge TDW, Schofield CJ. Angewandte Chemie Intl. Ed. (2013) 52(30):7709-13 [web][PDF] 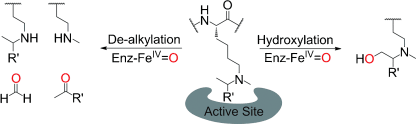 |
|
||||||||||||||||||||
(29) Selective Small Molecule Probes for the Hypoxia Inducible Factor (HIF) Prolyl Hydroxylases. Chowdhury R, Candela-Lena J, Chan M, Greenald D, Yeoh K, Tian Y, McDonough M, Tumber A, Rose NR, Conejo-Garcia A, Demetriades M, Mathavan S, Kawamura A, Lee MK, van Eeden F, Pugh C, Ratcliffe PJ, Schofield CJ. ACS Chemical Biology (2013) 8(7):1488-1496 19 [web][PDF]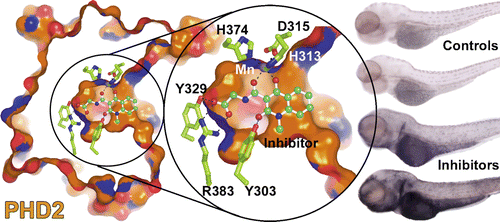 |
|
||||||||||||||||||||
(28) Dual-action inhibitors of HIF hydroxylases that induce binding of a second iron ion. Yeoh KK, Chan MC, Thalhammer A, Demetriades M, Chowdhury R, Tian YM, Stolze I, McNeill LA, Lee MK, Woon EC, Mackeen MM, Kawamura A, Ratcliffe PJ, Mecinović J, Schofield CJ. Org Biomol Chem (2013) 11 732-745 [web][PDF]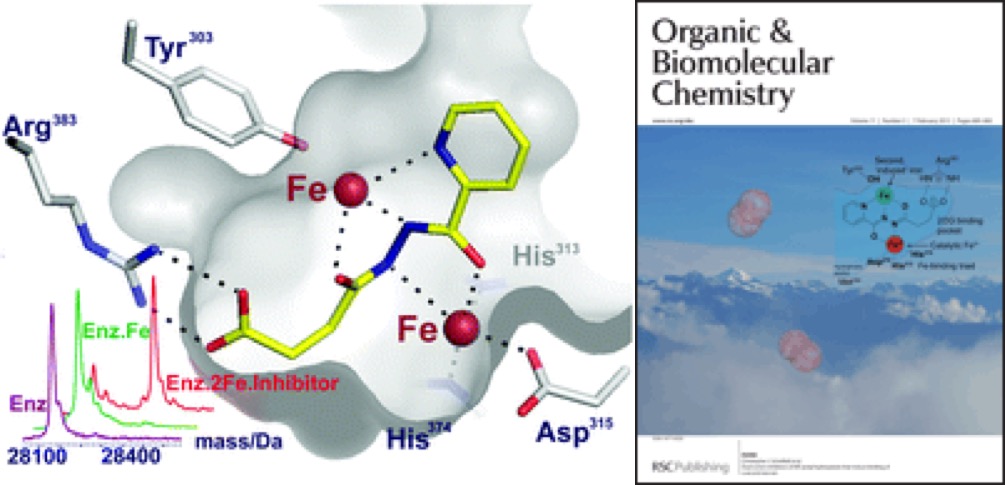 |
|
||||||||||||||||||||
2012 |
|||||||||||||||||||||
| (27) Piperidinols that show anti-tubercular activity as inhibitors of arylamine N-acetyltransferase: an essential enzyme for mycobacterial survival inside macrophages. Abuhammad A, Fullam E, Lowe ED, Staunton D, Kawamura A, Westwood IM, Bhakta S, Garner AC, Wilson DL, Seden PT, Davies SG, Russell AJ, Garman EF, Sim E. PloS ONE (2012) 7(12):e52790. [web][PDF] | |||||||||||||||||||||
(26) A reporter ligand NMR screening method for 2-oxogluarate inhibitors. Leung IK, Demetriades M, Hardy AP, Lejeune C, Smart TJ, Szollossi A, Kawamura A, Schofield CJ, Claridge TD. J. Med. Chem. (2012) 56(2):547-55 [web][PDF]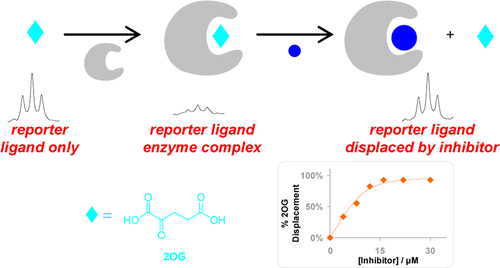 |
|
||||||||||||||||||||
(25) Plant Growth Regulator Daminozide Is a Selective Inhibitor of Human KDM2/7 Histone Demethylases. Rose NR, Woon EC, Tumber A, Walport LJ, Chowdhury R, Li XS, King ON, Lejeune C, Ng SS, Krojer T, Chan MC, Rydzik AM, Hopkinson RJ, Che KH, Daniel M, Strain-Damerell C, Gileadi C, Kochan G, Leung IK, Dunford J, Yeoh KK, Ratcliffe PJ, Burgess-Brown N, von Delft F, Muller S, Marsden B, Brennan PE, McDonough MA, Oppermann U, Klose RJ, Schofield CJ*, Kawamura A*. J. Med. Chem. (2012) 55(14) 6639-43. [web][PDF]. |
|
||||||||||||||||||||
(24) Linking of 2-Oxoglutarate and Substrate Binding Sites Enables Potent and Highly Selective Inhibition of JmjC Histone Demethylases. Woon, ECY, Tumber A, Kawamura A, Hillringhaus L, Ge W, Rose NR, Ma JHY, Chan MC, Walport LJ, Che KH, Ng SS, Marsden BD, Oppermann U, McDonough MA and Schofield CJ. Angewandte Chemie (2012) 51 (7) 1631-4 [web][PDF]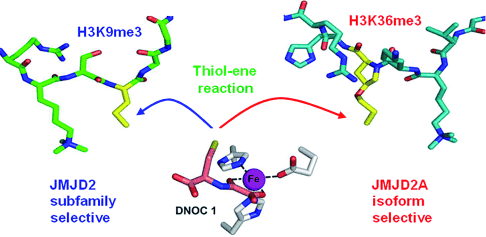 |
|
||||||||||||||||||||
2011 |
|||||||||||||||||||||
| (23) Mutations To Metabolic Enzymes In Cancer Herald A Need To Unify Genetics And Biochemistry. Kawamura A*, Loenarz C and Schofield CJ* Cell Cycle (2011) 10(17):2819-20 [web] | |||||||||||||||||||||
(22) A Photoreactive Small-Molecule Probe for 2-Oxoglutarate. Rotili D, Altun M, Kawamura A, Wolf A, Fischer R, Leung IKH, Mackeen MM, Tian Y, Ratcliffe PJ, Mai A, Kessler BM, Schofield CJ. Chem Biol (2011)18(5) 642-54 [web][PDF]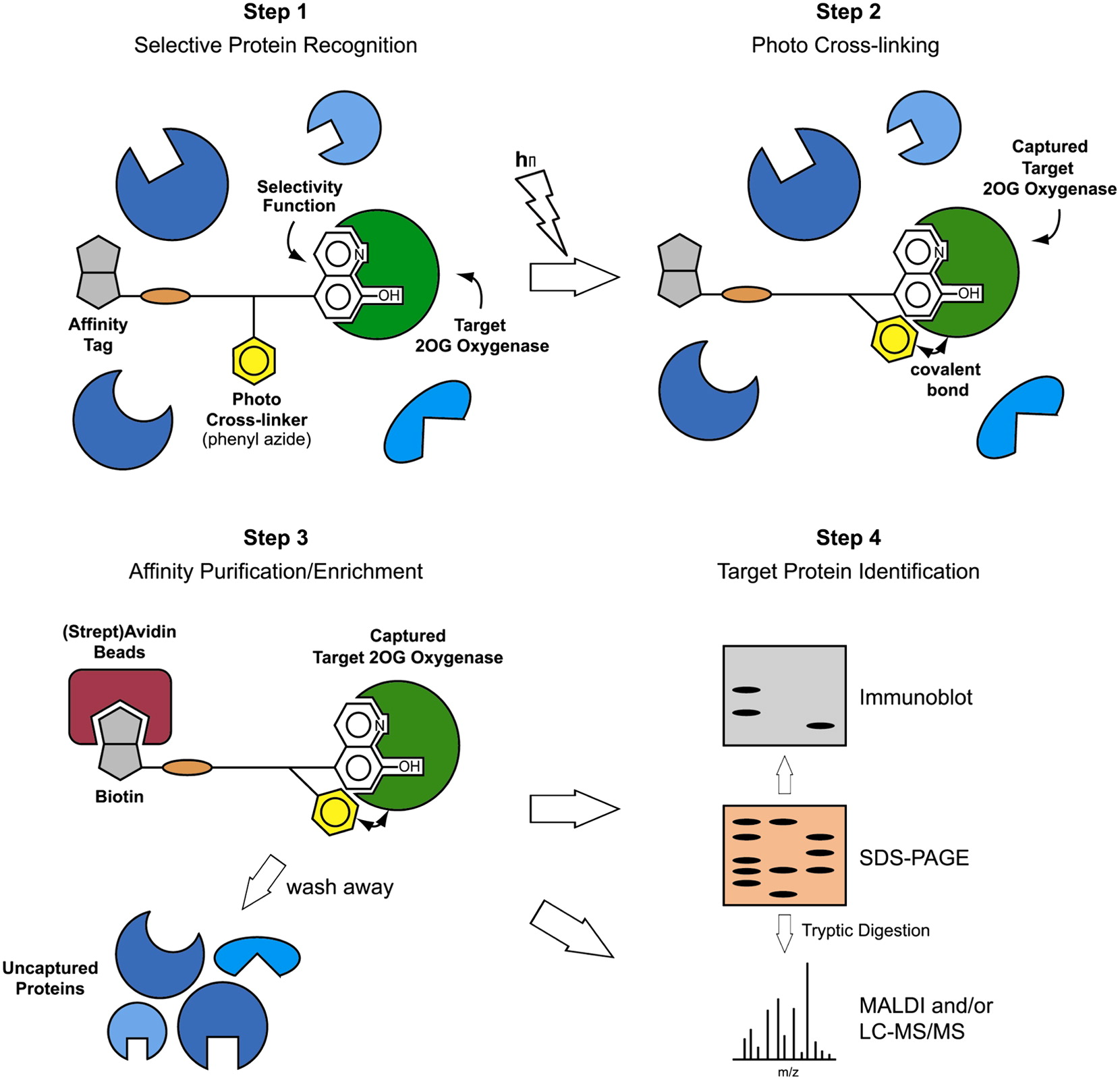 |
|
||||||||||||||||||||
| (21) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. Chowdhury R, Yeoh K, Tian Y, Hillringhaus L, Bagg E, Rose NR, Leung IK, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ* and Kawamura A* EMBO Reports (2011) 12(5): 463-9 [web][PDF] | |||||||||||||||||||||
(20) Inhibition of 2-Oxoglutarate Dependent Oxygenases. Rose NR, McDonough MA, King OF, Kawamura A, Schofield CJ. Chemical Society Reviews (2011) 40(8):4364-9 [web][PDF] |
|
||||||||||||||||||||
2010 |
|||||||||||||||||||||
| (18) Quantitative High-Throughput Screening Identifies 8-Hydroxyquinolines as Cell-Active Histone Demethylase Inhibitors. King ON, Li XS, Sakurai M, Kawamura A, Rose NR, Ng SN, Quinn AM, Rai G, Mott BT, Klose RJ, Oppermann U, Heightman T, Maloney DJ, Jadhav A, Schofield CJ, Simeonov A. PLoS One (2010) 5(11): e15535 [web][PDF] | |||||||||||||||||||||
| (17) Development of homogeneous luminescence assays for histone demethylase catalysis and binding. Kawamura A, Tumber A, Rose NR, King ON, Daniel M, Oppermann U, Heightman TD, Schofield CJ. Anal Biochem (2010) 404 86 [web][PDF] | |||||||||||||||||||||
(16) Small molecule colorimetric probes for specific detection of human arylamine N-acetyltransferase 1, a potential breast cancer biomarker. Laurieri N, Crawford MH, Kawamura A, Westwood IM, Robinson J, Fletcher AM, Davies SG, Sim E, Russell AJ. J Am Chem Soc. (2010) 132(10):3238-9 [web][PDF]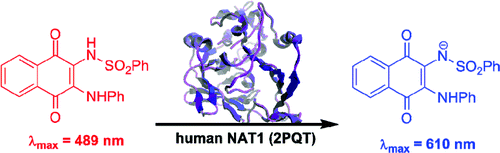 |
|
||||||||||||||||||||
| (15) Identification of arylamine N-acetyltransferase inhibitors as an approach towards novel anti-tuberculars. Westwood I, Bhakta S, Russell AJ, Fullam E, Anderton MC, Kawamura A, Mulvaney AW, Vickers R.J, Bhowruth V, Besra G.S, Lalvani A, Davies S.G, and Sim E. Protein & Cell (2010) 1 (1) 82-95 [web][PDF] | |||||||||||||||||||||
2002-2009 |
|||||||||||||||||||||
| (14) Comparison of the Arylamine N-acetyltransferase from Mycobacterium marinum and Mycobacterium tuberculosis. Fullam E, Kawamura A, Wilkinson H, Abuhammad A, Westwood I, Sim E. Protein J. (2009) 28(6):281-93 [web] | |||||||||||||||||||||
| (13) Selective small molecule inhibitors of the potential breast cancer marker, human arylamine N-acetyltransferase 1, and its murine homologue, mouse arylamine N-acetyltransferase 2. Russell AJ, Westwood IM, Crawford MH, Robinson J, Kawamura A, Redfield C, Laurieri N, Lowe ED, Davies SG, Sim E. Bioorg Med Chem (2009) 17(2) 905-18 [web] | |||||||||||||||||||||
| (12) Temperature stability of proteins essential for the intracellular survival of Mycobacterium tuberculosis. Lack NA, Kawamura A, Fullam E, Laurieri N, Beard S, Russell AJ, Evangelopoulos D, Westwood I, Sim E. Biochem J. (2009) 418(2) 369-78 [web] | |||||||||||||||||||||
| (11) Mouse N-acetyltransferase type 2, the homologue of human N-acetyltransferase type 1. Kawamura A, Westwood, I., Wakefield, L., Long, H., Zhang, N., Walters, K., Redfield, C and Sim, E. Biochem Pharm (2008) 75 (7) 868-878 [web] | |||||||||||||||||||||
| (10) Arylamine N-acetyltransferases: Structural and Functional Implications of Polymorphisms. Sim E, Lack N, Wang C, Long H, Westwood I, Fullam E and Kawamura A. Toxicology (2008) 254(4) 170-83 [web] | |||||||||||||||||||||
| (9) Synthesis, metabolic stability, and anti-cancer activities of novel amonafide derivatives. Norton JT, Witschi MA, Luong L, Kawamura A, Ghosh S, Stack MS, Sim E, Avram MJ, Appella DH and Huang A. Anti-Cancer Drugs (2008) 19 (1) 23-36 [web] | |||||||||||||||||||||
| (8) Mouse arylamine N-acetyltransferase 2 (Nat2) expression in the developing neuroendocrine system. Wakefield L, Cornish V, Long H, Kawamura A, Zhang X, Hein DW and Sim E. Biomarkers (2007) Sept 25; 1-13 [web] | |||||||||||||||||||||
| (7) Description of a novel polymorphic gene encoding for arylamine N-acetyltransferase in the rhesus macaque (Macaca mulatta), a model animal for endometriosis. Fakis G, Boukouvala S, Kawamura A, and Kennedy S. Pharmacogenetics and Genomics (2007) 17 (3) 181-188 [web] | |||||||||||||||||||||
| (6) Structure and mechanism of arylamine N- acetyltransferases. Westwood IM, Kawamura A, Fullam E, Russell AJ, Davies SG, and Sim E Current Topics in Medicinal Chemistry (2006) 6 1641-1654 [web] | |||||||||||||||||||||
| (5) Eukaryotic arylamine N-acetyltransferase: Investigation of substrate specificity by high-throughput screening. Kawamura A, Graham J, Mushtaq A, Vath GM, Hanna PE, Wagner CR and Sim E. Biochem Pharm (2005) 69 347-359 [web] | |||||||||||||||||||||
| (4) Overexpression, Purification, and Characterization of Recombinant Human Arylamine N-Acetyltransferase 1. Wang H, Vath GM, Kawamura A, Bates CA, Sim E, Hanna PE, Wagner CR. Protein J (2005) 2 65-77 [web] | |||||||||||||||||||||
| (3) Structural investigation of mutant Mycobacterium smegmatis arylamine N-acetyltransferase: a model for a naturally occurring functional polymorphism in Mycobacterium tuberculosis arylamine N-acetyltransferase. Kawamura A, Sandy J, Upton A, Noble M and Sim E. Prot Exp Purif (2003) 27 75-84 [web] | |||||||||||||||||||||
| (2) The structure of arylamine N-acetyltransferase from Mycobacterium smegmatis – an enzyme which inactivates the anti-tubercular drug, isoniazid. Sandy J, Mushtaq A, Kawamura A, Sinclair J, Sim E and Noble M. J Mol Biol (2002) 318 1071-1083 [web] | |||||||||||||||||||||
| (1) The pharmacogenetics of NAT: structural aspects. Pompeo F, Brooke E, Kawamura A, Mustaq A, and Sim E. Pharmacogenomics (2002) 3 (1) 19-30 [web] |