Our research focuses on understanding the molecular mechanisms of biological processes. We employ a variety of biochemical, biophysical and cell-based functional studies of proteins, alongside development of novel cyclic peptide and small molecule based chemical tools. We have a particular interest in proteins that regulate epigenetic modifications on histones and DNA, and our work seeks to improve understanding of their mechanisms and how they influence the expression of genes. We also tackle challenging protein-protein interactions of biomedical importance, including chemokine signalling and inflammation. We work at the interface of chemistry, biology and medicine, and many of our projects are highly collaborative.

Areas of Interest
Chemical Epigenetics
Epigenetic modifications refer to the covalent modifications to DNA and histones which constitute a set of inherited changes in gene expression, without changing the underlying DNA sequences. By having a molecular understanding and developing chemical probes against the proteins that regulate these epigenetic modifications, we hope to gain a better understanding of epigenetic mechanisms and how they influence the expression of genes in health and disease.
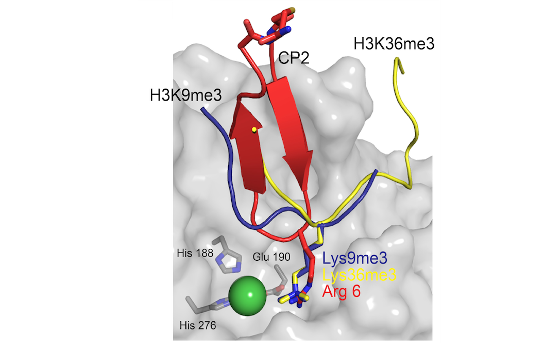
Histone modifiers: Abnormal histone post-translational modification (PTM) patterns and dysregulation of epigenetic enzymes are implicated in many diseases, including cancer. We study families of epigenetic enzymes that are ‘erasers’ and ‘writers’ of histone PTMs (e.g. histone demethylases (KDM) and methyltransferases). Many of these proteins also contain ‘reader’ domains (e.g. PHD-fingers) which allow them to recognise specific epigenetic marks. Our work has led to the development of new assay platforms, new functional and mechanistic insights into epigenetic enzymes and proteins, factors affecting epigenetic regulation (e.g. O2, (onco)metabolite levels) as well as generation of chemical tools/probes (Figure 1).
Current projects include:
- Biochemical and cellular studies of histone demethylases (CRUK Programme Grant)
- Investigating the inter-domain and combinatorial PTM cross-talk
- Molecular recognition of PTMs by epigenetic proteins
- Chemical probe development for epigenetic proteins including KDMs and PHD-finger domains (ERC EPITOOLS)
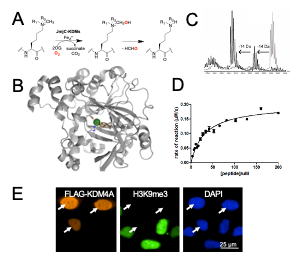
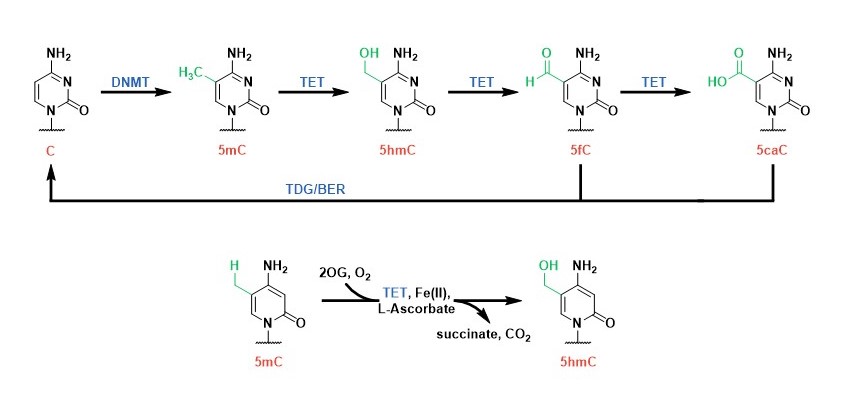
DNA Modifiers: Cytosine methylation is established by DNA methyltransferases (DNMTs) and is an important epigenetic mark that regulate gene expression. We study a family of 2OG oxygenases called Ten-Eleven Translocation enzymes (or TETs) which play a key role in the cytosine modification cycle by oxidising methylated cytosines (Fig 2). We have developed activity- and binding-based biochemical assays to study these enzymes. We have also identified potent TET cyclic peptide inhibitors and are working to develop chemical probes which would enable biological role investigations in a cellular context.
Inflammation
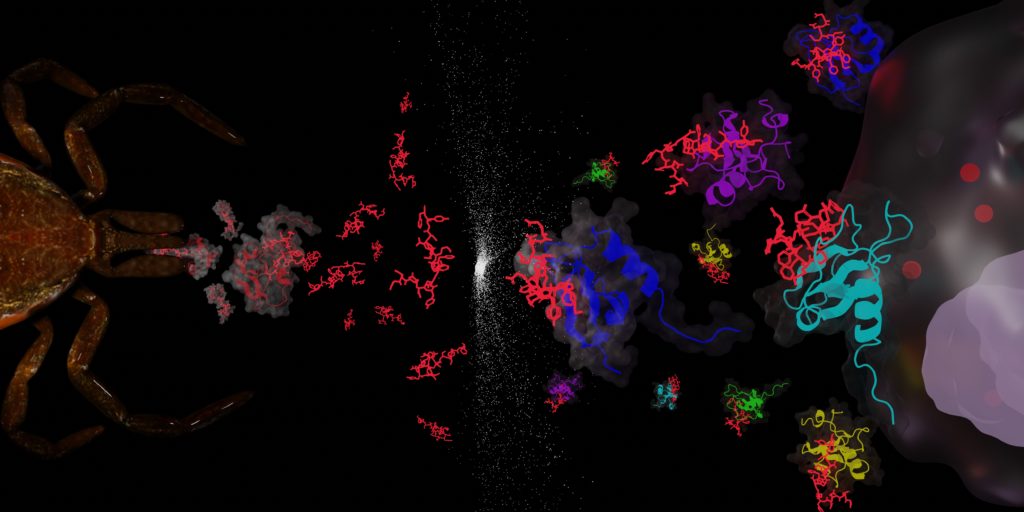
Inflammation processes are at the core of many pathologies including several major cardiovascular diseases such as myocarditis, atherosclerosis, and strokes, and influence the fate and drug resistance of cancer. Therefore, leukocyte mobilisation and actions are responsible for many fatalities worldwide and have been the focus of many pharmaceutical development in the last 50 years. Our team is interested in targeting the chemotactic cytokine (chemokine) network responsible for the leukocyte tissue infiltration using tick saliva proteins (evasins) and peptides inspired from these proteins (Figure 3).
Peptide Discovery Platform
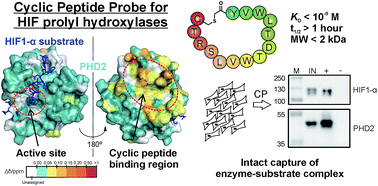
We utilise a combination of peptide chemistry and peptide-array / mRNA-display technologies for substrate profiling, protein-protein interaction mapping, as well as to generate highly potent and selective ligands against our targets of interest. Some highlights in this area include: substrate profiling of epigenetic enzymes, development of potent and selective cyclic peptides inhibitors against epigenetic enzymes (Figure 4), as well as affinity cyclic peptide probes for oxygen sensing enzyme PHD2 (Figure 5).